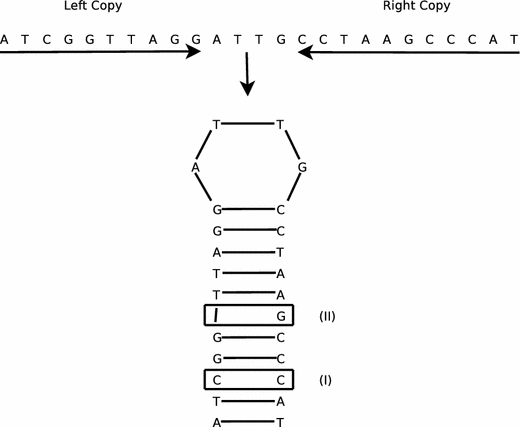
![Comparative plastomics of Amaryllidaceae: inverted repeat expansion and the degradation of the ndh genes in Strumaria truncata Jacq. [PeerJ] Comparative plastomics of Amaryllidaceae: inverted repeat expansion and the degradation of the ndh genes in Strumaria truncata Jacq. [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/12400/1/fig-1-full.png)
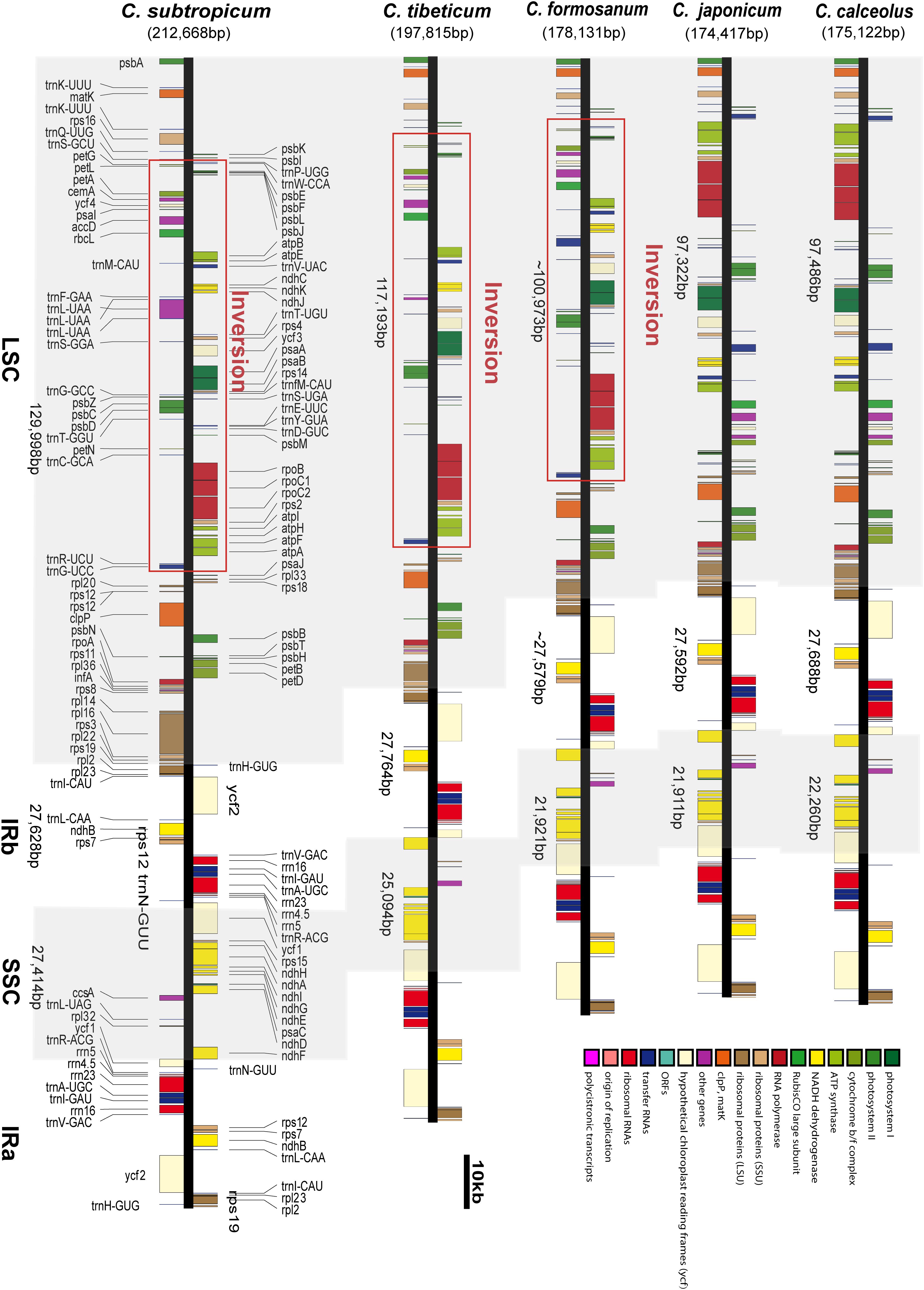
Comparative plastomics of Amaryllidaceae: inverted repeat expansion and the degradation of the ndh genes in Strumaria truncata Jacq. [PeerJ]
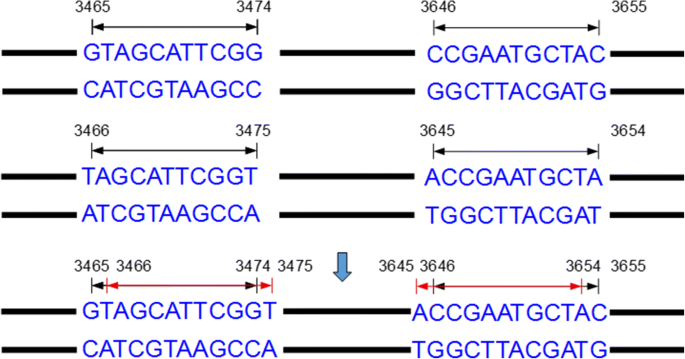
MiteFinderII: a novel tool to identify miniature inverted-repeat transposable elements hidden in eukaryotic genomes | BMC Medical Genomics | Full Text
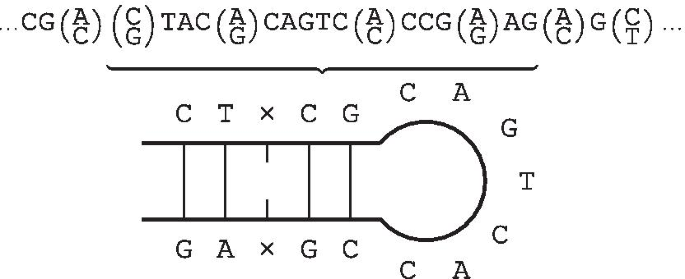
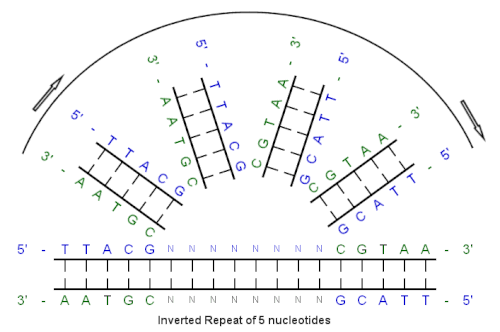
IUPACpal: efficient identification of inverted repeats in IUPAC-encoded DNA sequences | BMC Bioinformatics | Full Text
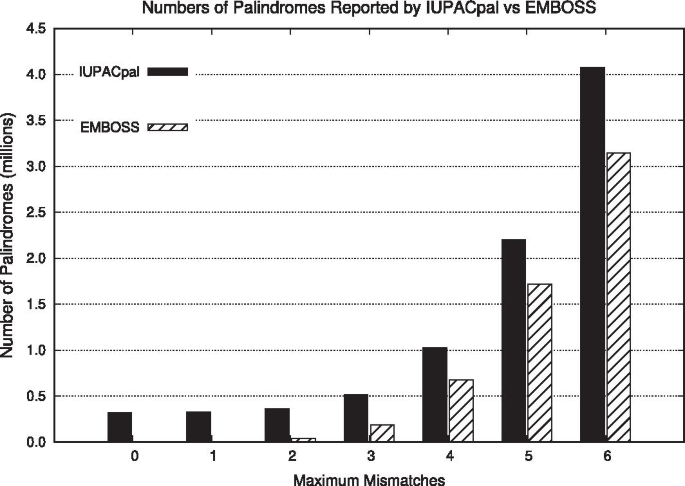
IUPACpal: efficient identification of inverted repeats in IUPAC-encoded DNA sequences | BMC Bioinformatics | Full Text
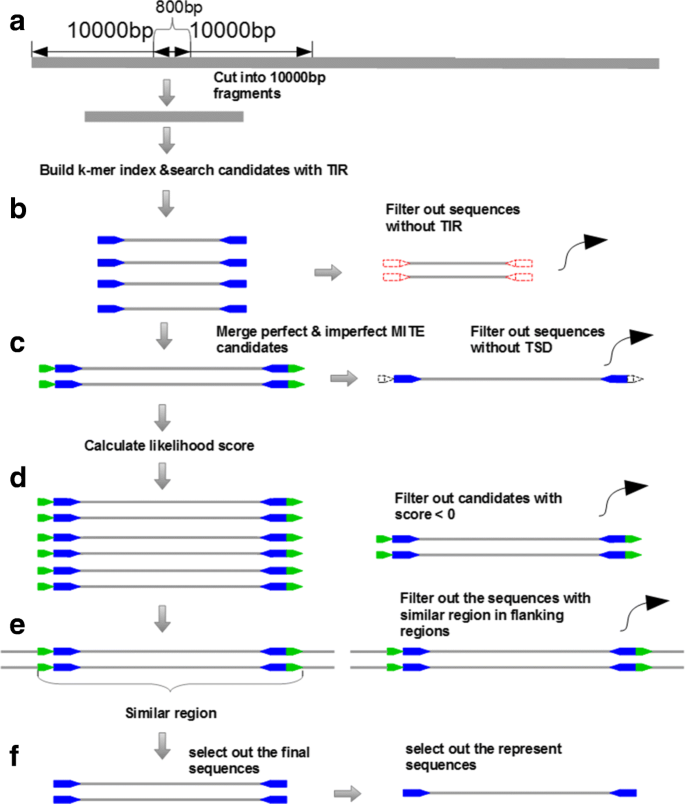
MiteFinderII: a novel tool to identify miniature inverted-repeat transposable elements hidden in eukaryotic genomes | BMC Medical Genomics | Full Text

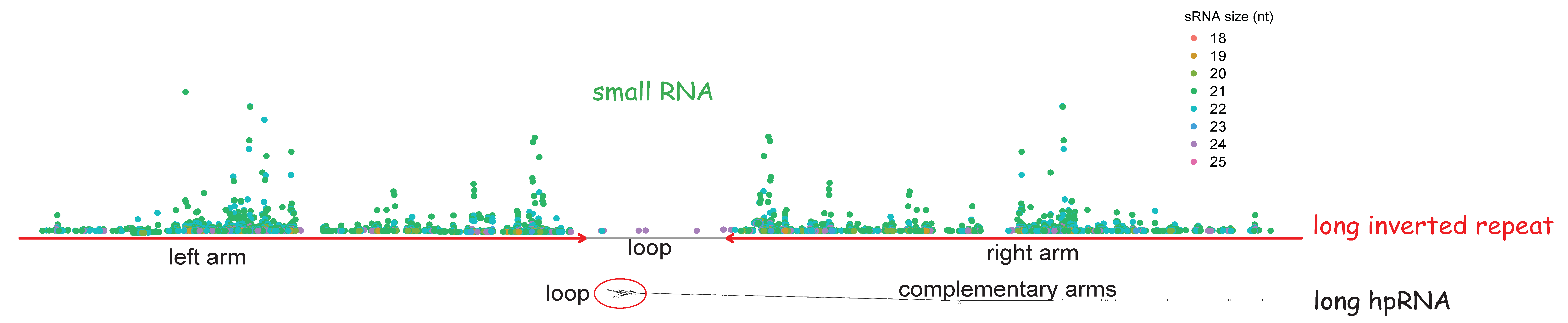
Correlations between long inverted repeat (LIR) features, deletion size and distance from breakpoint in human gross gene deletions | Scientific Reports

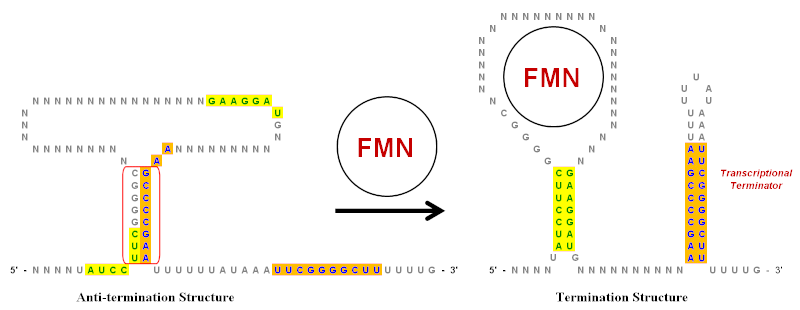
Global analysis of inverted repeat sequences in human gene promoters reveals their non-random distribution and association with specific biological pathways - ScienceDirect